👋 Hello there, I’m Tommaso
📈 I’m interested in data analysis and machine learning problems.
👨💻 I develop software instruments and methods that biology experts can use to improve our understanding of current and future viral threats.
🧬🦠 Doing bioinformatic is my way of trying to make the world a better place.
🔍 Research areas in Information Technology
Anomaly detection
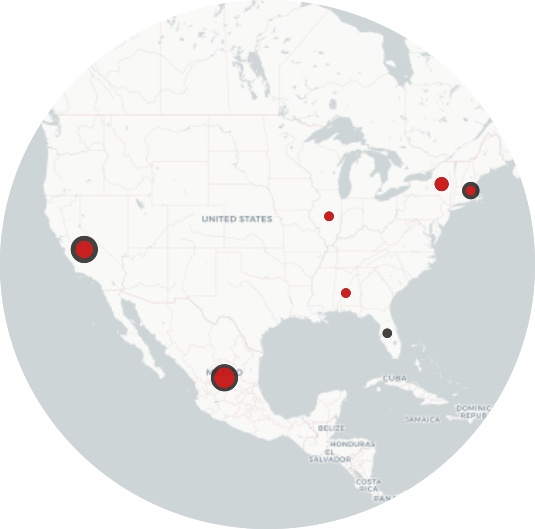 | Application field: Divergent viral evolution By integrating multidimensional anomaly detection algorithms with codon bias and dinucleotide composition, I developed a method to detect viral isolates featuring a high zoonotic risk. This method can be used to enhance genomic surveillance practices and, possibly, to anticipate pandemics. |
Statistical methods
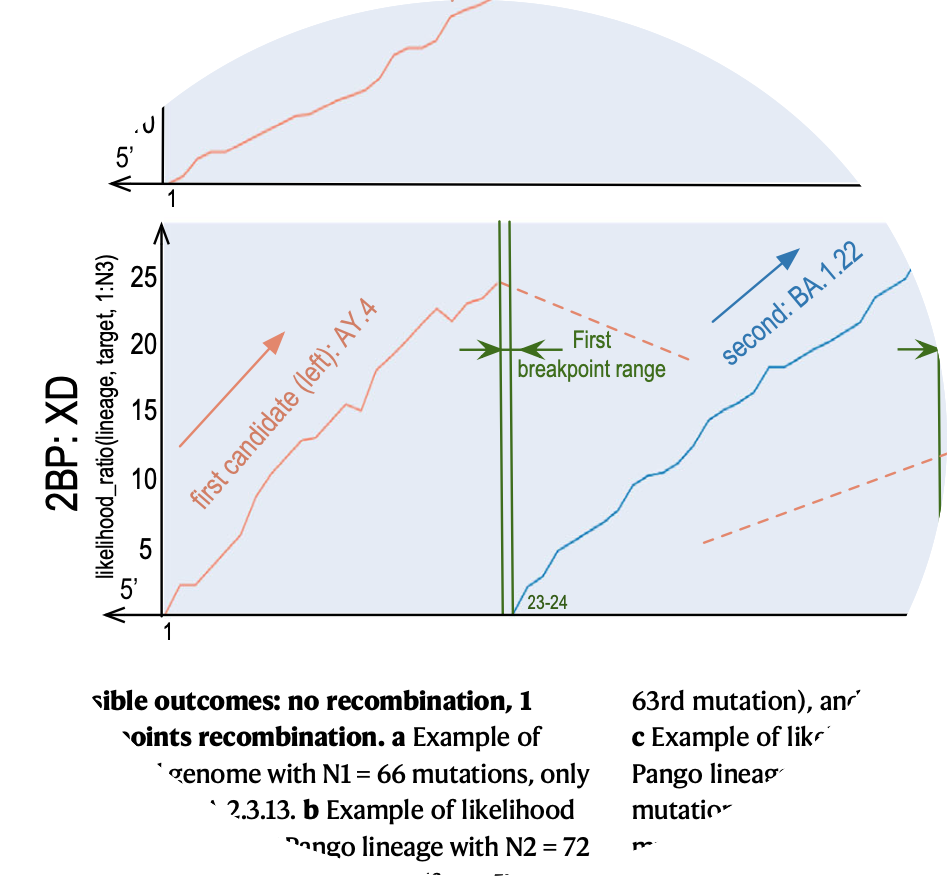 | Application field: Viral recombination Using robust statistical methods, I developed a cutting-edge method that significantly heightens the detection rate and speed of recombination events, offering a vital asset to combat future pandemics or epidemics. (link) |
Machine Learning
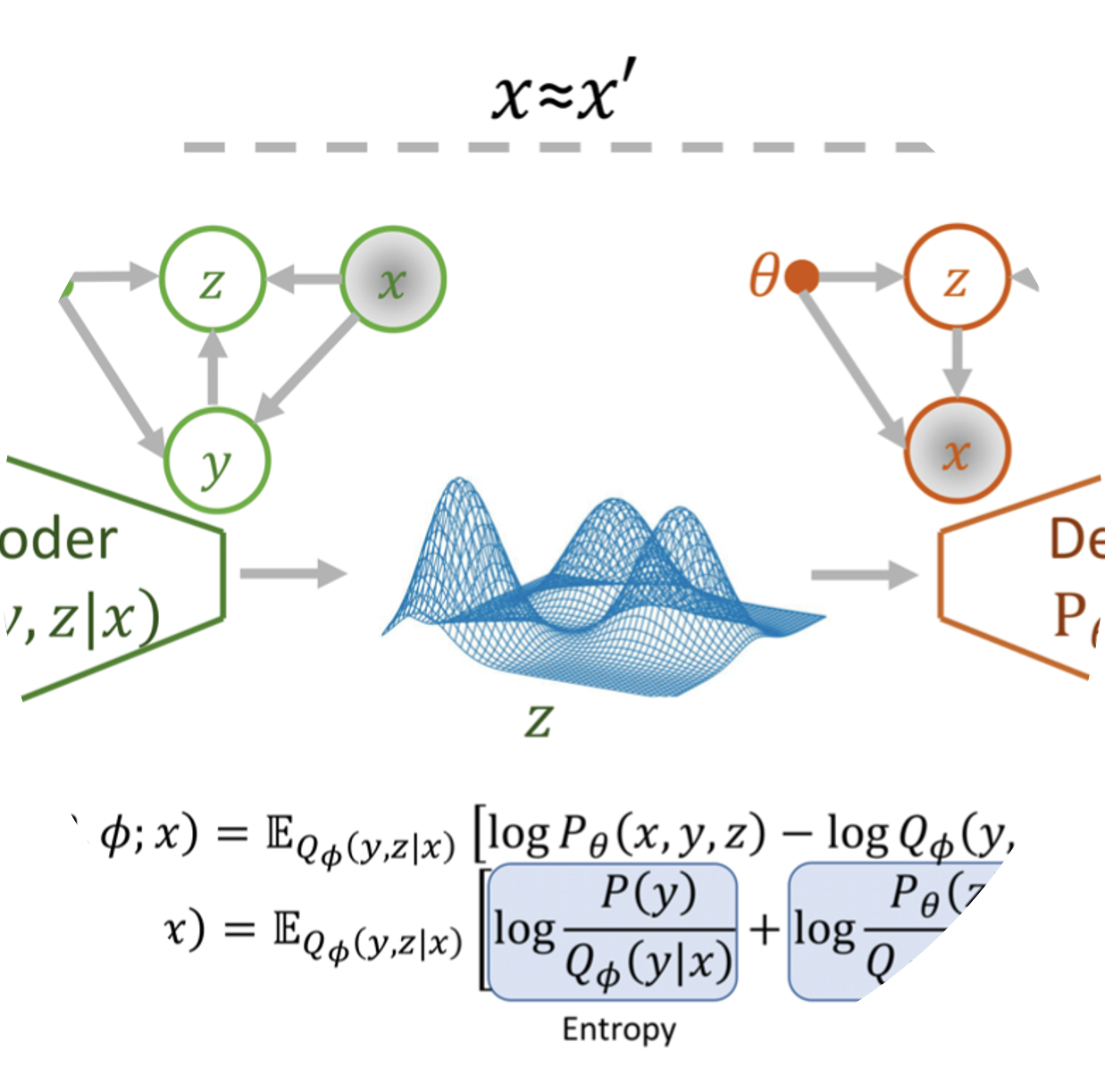 | Application field: Clustering of viruses Using information compression and clustering algorithms on viral sequences it is possible to highlight key epidemiological events and enable detailed comparisons of genomic features across Influenza A Virus serotypes. This approach provides a scalable framework to study viral evolution, adaptation, especially in viruses with limited genomic data available. (link) Alternative and completely unsupervised methods, like Gaussian Mixture Variational AutoEncoders (GMVAE) have also been explored on few selected cases. |
Knowledge Integration and Management
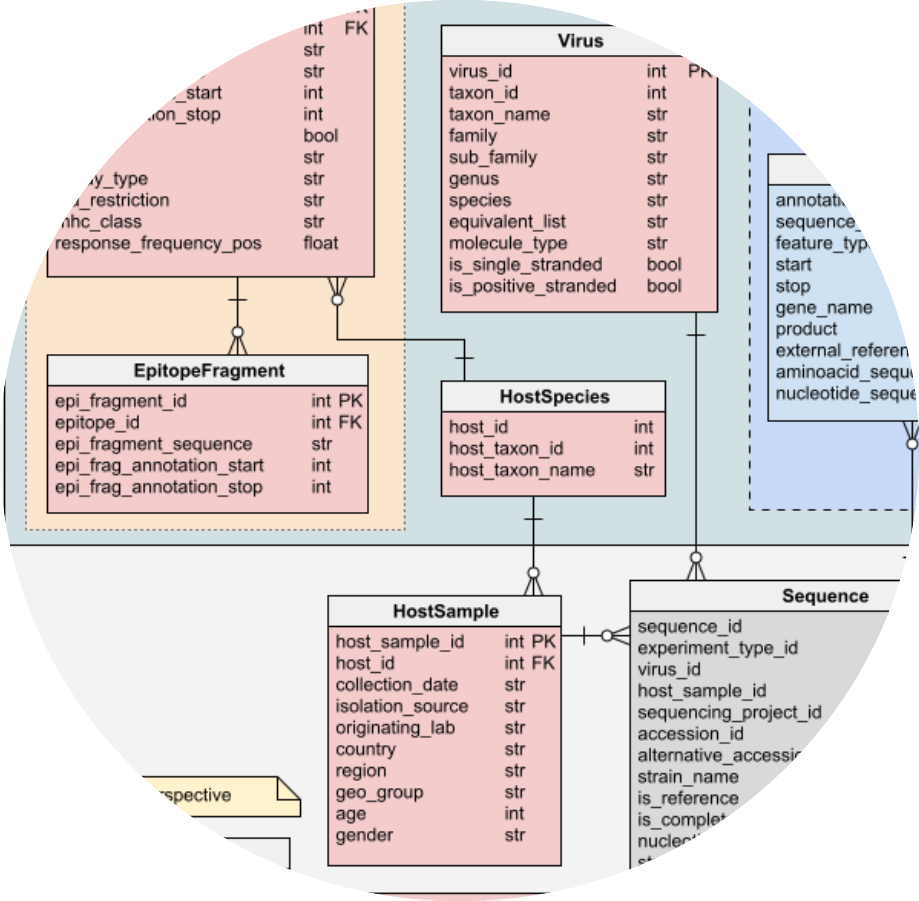 | Application field: Data modeling & integration pipelines Fast, reliable, incremental integration of viral sequences to develop one of the largest relational databases of viral genomes in 2020 (ViruSurf). I also contributed to crafting tools facilitate the analysis of SARS-CoV- 2 virus evolution (VirusViz, EpiSurf, CoV2K). Application field: AI reasoning on knowledge graphs I used a knowledge and reasoning representation language to bring artificial intelligence to a knowledge-base (CoV2K), answering interesting questions about SARS-CoV-2 variants and phenotypic effcts. (link) |
Population Genomics
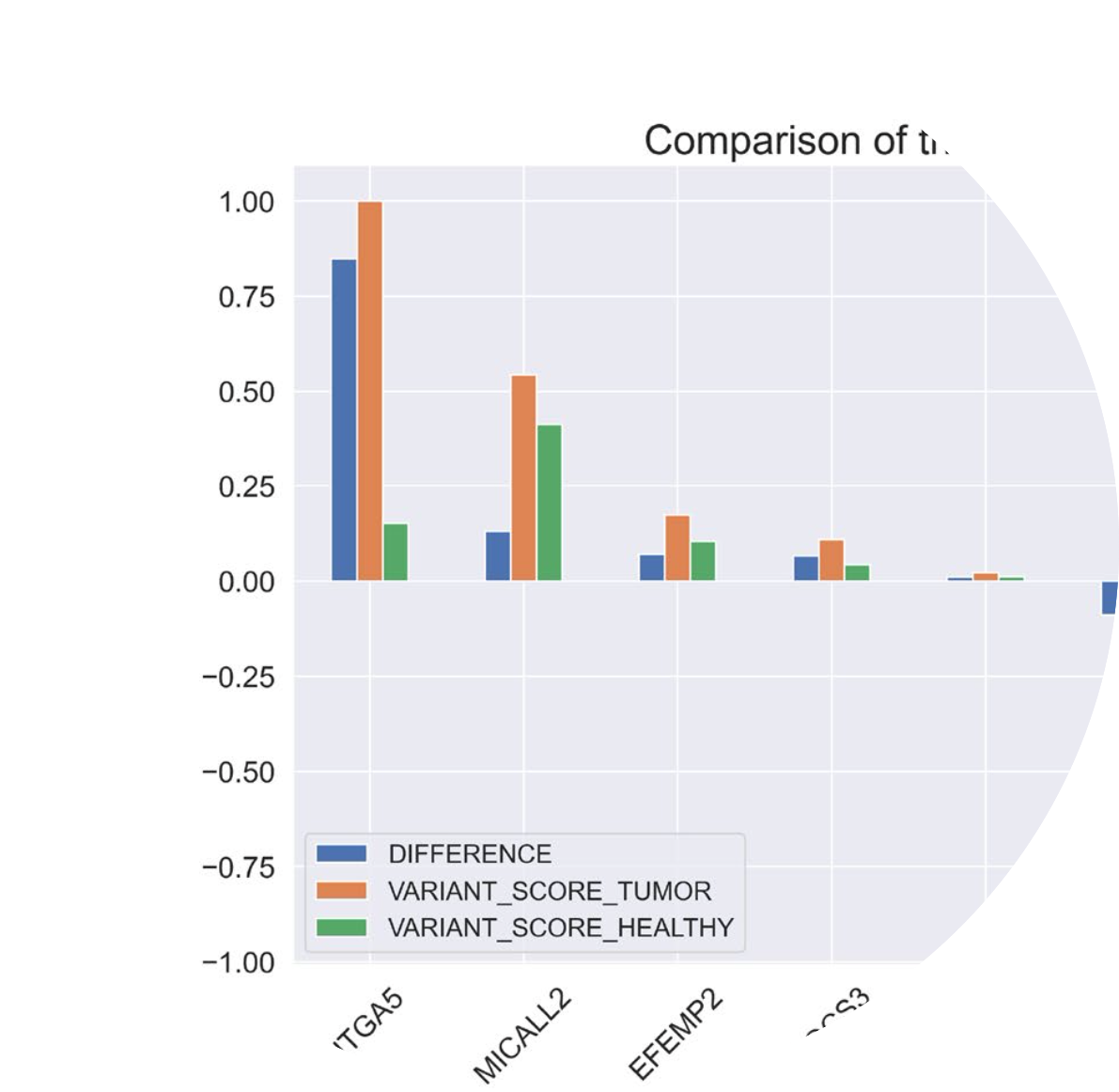 | Application field: Control population studies, cancer target gene identification I built an API to ease the selection of genomic populations and assess genomic characteristcs in the 1000 Genomes Project, and The Cancer Genome Atlas (TCGA). Combining gene annotations, this API provides summary information about any population defined with fine-grained filters on DNA variants and metadata. (link) |
🎓 Education and Training
- Postdoctoral Research Fellow
- PhD cum laude in Information Technology @ Politecnico di Milano, Italy
- Artificial Intelligence and Machine Learning course @ University of Oxford, UK
- Master Degree in Computer Science and Engineering @ Politecnico di Milano, Italy